Computer Vision and Imaging Extended [06 30241]
Assessed Assignment
B15 2TT
February 2024
1. Instructions
You are to work individually for this assessment, and produce one report. The report must be no more than THREE pages long including graphs and tables. The report should be submitted via CANVAS (See Canvas for deadline).
Your assignment involves some implementation and some experimentation, plus the write up. All images to be processed are provided.
Your code/script should also be submitted as supplementary data (5 extra Pages max): Remember to follow good practice with structure, use of functions and adding of comments to your code.
Any text beyond the stated 3 pages will not be marked. There will be 5% penalty per day (or part of), for late submissions.
Your submitted assignment should:
. Be a maximum of 3 pages (A4)
. Have minimum margins (top/bottom/sides) of 2 cm
. Use Arial (narrow) font of 11
. Contain your student ID, and module code in the header
2. The assignment:
Often, given a set of images, we are required to segment out regions/labels before further analysis of our data. For this task, you are given 10 consecutive ‘axial’ cross-sections of a publicly available MRI of a single human subject and are required to segment out 5 tissue layers. Specifically, you are provided a single matlab file, ‘Brain.mat’ which contains T1 weighted MRI images (each slice has a dimension of 362*434 pixel) with each z-slice being at 1 mm interval (spanning 10 mm), Figure 1.
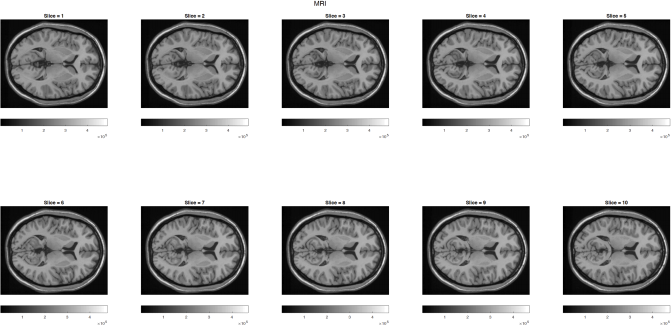
Figure 1: T1 weighted MRI data at 10 consecutive slices
Additionally you are given a ‘pre-segmented’ set of the same data to allow comparison and analysis, Figure 2.
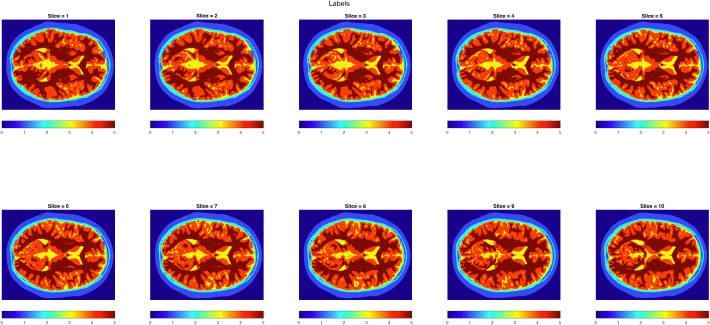
Figure 2: Segmented ‘labelled’ regions of the T1 weighted data in Figure 1
The region labels used in Figure 2 are:
. Label 0 = air
. Label 1 = skin/scalp
. Label 2 = skull
. Label 3 = CSF
. Label 4 = Gray Matter
. Label 5 = White Matter
Loading .mat files in python:
. from scipy.io import loadmat
. import sys
. data = loadmat('Brain.mat')
. T1 = data['T1']
. label = data['label']
Task 1: 2D tissue segmentation [20 Marks]
. Develop and apply different segmentation algorithms, based on any technique you have learnt to each slice of the MRI data. You need to apply exactly the same algorithm to every slice. HINT: This will more than likely be a combination of different techniques.
Task 2: Result evaluation [10 Marks]
. Compare your segmented results for each algorithm to the ground-truth label provided. Justify and explain the metric used to assess accuracy. Based on your evaluation and results, highlight the best algorithm to be used.
Task 3: Advanced 2D tissue segmentation [20 Marks]
. Think about and implement a 3D image segmentation algorithm that can be applied to all slices, simultaneously. Discuss why the use of the proposed algorithm is better/worse than those in Task 1 by utilizing the same evaluation metric applied in Task 2.
Write an experimental report of THREE pages detailing the experiments you have carried out. Make sure that you include the Aim, Method, Results and Conclusions. You should be able to draw a limited conclusion about which algorithm or algorithms are best.
Notes
. Remember, credit will be given where there is detail and reasoning. If you use any algorithm NOT used in class, you need to outline its implementation.
. Although I have made some possible suggestions about how to analyse each technique you are free to do your own investigations and report them. You will obtain significant credit for doing this if your investigations are interesting and well reported. You will also gain considerable credit for extending the techniques in other ways.