Hello, if you have any need, please feel free to consult us, this is my wechat: wx91due
Semester 1, 2024 CITS4402 Computer Vision
Project: Applications of Computer Vision in Medicine: Magnetic Resonance Imaging Classification for Glioma Diagnosis
Timeline
Week 7 to Week 12: Work on your project. You have 6 weeks. The facilitators will be available in the labs during the lab hours for questions.
Week 12 (21st & 22nd of May 2024): Presentation of the projects during the lab hours. The schedule will be announced.
Due: Friday, 24th of May 2024, 4 PM (NO EXTENSIONS)
Grouping
Form groups of 3 students and include the first names and surnames of the group members on LMS Discussion board. This should have been done by now.
Applications of Computer Vision in Medicine: Magnetic Resonance Imaging Classification for Glioma Diagnosis
Background
Medical imaging is a critical application of Computer Vision (CV). In this project, we will apply CV techniques to Magnetic Resonance Imaging (MRI) for glioma diagnosis. Glioma is a type of malignant brain tumor with varying degrees of severity. Gliomas can be broadly categorized into Low-Grade Gliomas (LGG) and High-Grade Gliomas (HGG), with LGG being less aggressive and HGG being more aggressive. Accurate diagnosis is crucial for clinical decision-making as the intervention and prognosis significantly differ between LGG and HGG patients. Currently, the diagnosis involves both (i) non- invasive imaging studies and (ii) invasive histopathological examinations, with the latter being essential for a definitive diagnosis. However, invasive examinations, such as biopsy or surgical resection of the tumor tissue, pose high risks to patients. Therefore, there is a pressing need to perform glioma diagnosis solely based on non-invasive imaging studies.
A commonly used medical imaging modality for non-invasive imaging studies is MRI. MRI uses strong magnetic fields and radio waves to construct volumetric images of the internal structures of the body. It is used for imaging patients with glioma due to its ability to provide excellent contrast between brain tumor and normal brain tissues. Glioma exhibits extreme heterogeneity in appearance and shape when visualized on MRI, making image-based diagnosis with the naked eye very challenging. However, LGG/HGG may have certain intrinsic, unique imaging features, suggesting the potential for leveraging CV techniques for image-based diagnosis.
Project Overview
In this project, our objective is to use CV techniques to classify patients with LGG or HGG based on their MRI studies. The project can be divided into the following steps.
• Step One: Data sourcing;
• Step Two: Visualization;
• Step Three: Feature Detection and Extraction;
• Step Four: Feature Selection;
• Step Five: Classification using SVM.
Step One: Data Sourcing
The dataset we are using is publicly available on Kaggle. You need to first register on Kaggle and download the dataset.
Source of data:https://www.kaggle.com/datasets/awsaf49/brats2020-training-data/data
The dataset contains MRI of 369 patients with glioma. An MRI volume is a volumetric image comprising n slices (layers of the volume). Each slice comprises h × w pixels; for the entire volume, there are h × w × nvoxels (pixels in 3D). In this dataset, n=155 for every MRI volume; the MRI slices are stored as H5 files. The filenames follow the pattern volume_[volume ID]_slice_[slice ID], e.g., volume 1 slice_0 suggests the 0th slice of the 1st volume. Each H5 file contains four h × w images and three segmentation masks for three tumor sub-regions. The three tumor sub-regions include (i) the necrotic tumor core, (ii) the non-necrotic tumor core, and (iii) the surrounding tissues invaded by the tumor. For simplicity, the three segmentation masks can be merged to represent the whole tumor; however, you are free to perform an analysis for each sub-region.
The four images detail the same anatomical position and differ in terms of image acquisition protocol. They can be considered as four different channels (sequences), similar to the R, G, B channels of a natural image; however, for subsequent operation, they have to be processed separately. The four images are stored inah × w × 4 array: the 1st, 2nd, 3rd, 4th (MATLAB)/0th, 1st, 2nd, 3rd (Python) ‘layer’ of the array corresponds to the T2 Fluid Attenuated Inversion Recovery (T2- FLAIR), native (T1), post-contrast T1-weighted (T1Gd), T2-weighted (T2) channel, respectively. An example of the four channels of the same anatomical position is shown in Fig. 1. While the knowledge of MRI is out of scope for this course, and not needed for this project, interested readers may refer to the following resources:
• https://en.wikipedia.org/wiki/Magnetic_resonance_imaging#Sequences
• https://rads.web.unc.edu/wp-content/uploads/sites/12234/2018/05/Phy-MRI-Made- Easy.pdf
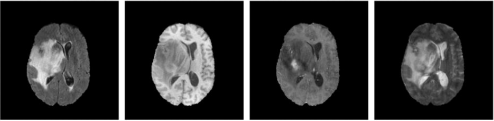
Figure 1: The four channels of the same anatomical position. Left to right: T2-FLAIR, T1, T1Gd, T2.
The name_mapping.xlsx file contains information regarding the grading of the patients with glioma from whom the MRI was acquired. Column A contains the diagnosis information; column F contains the corresponding volume ID.
Step Two: Visualization
In radiology departments, specialized software is often used for visualization, allowing clinicians to easily examine and annotate MRI volumes in a slice-by-slice manner. Some open-source software includes 3D Slicer (https://www.slicer.org/) and ITK-SNAP (http://www.itksnap.org/). Feel free to download one of the software and explore it.
For visualization, the task is to utilize the MATLAB/Python GUI to build a visualization tool mimicking the specialized software. The visualization tool should allow selecting a directory containing multiple H5 files, selecting the MRI channel to be displayed, turning on/off the tumor mask superimposed on the MRI image, and browsing through the MRI slices. Detailed requirements for the GUI can be found in the Deliverables section.
Step Three: Feature Detection and Extraction
There are two types of features that we are going to explore: (i) conventional features and (ii) radiomic features.
Conventional Features
Conventional features are more interpretable to human and are often recognized as imaging biomarkers for clinical decision-making. In the context imaging glioma, conventional features can be further divided into two subgroups: features encoding tumor (i) size and (ii) location information. Three conventional features are required for each MRI volume: (1) Maximum Tumor Area (encoding size information); (2) Maximum Tumor Diameter (encoding size information); (3) Outer Layer Involvement (encoding location information).
1. Maximum Tumor Area
On a slice, the tumor area is defined as the number of tumorous pixels. For all slices of a given volume, calculate the Maximum Tumor Area.
2. Maximum Tumor Diameter
On a slice, the tumor diameter is defined as the longest linear measurement (pixels) across the largest tumor component on the slice; the longest linear measurement across any component can be measured using principal component analysis of the component.
For all slices of a given volume, calculate the Maximum Tumor Diameter.
3. Outer Layer Involvement
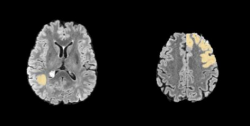
The outer layer of the brain is one of the most important regions, as it is where the cerebral cortex, which is responsible for cognition, is located. Hence, the outer layer involvement is a key indicator of the likelihood of cognitive disorder. A pair of examples of the outer layer not invaded/invaded by the brain tumor is shown in Fig. 2.
Assume that on all slices, the outer layer of the brain has a constant thickness of 5 pixels. For a given volume, calculate the percentage of Outer Layer Involvement.
Figure 2 Left: the outer layer not invaded by the brain tumor. Right: the outer layer invaded by the brain tumor.
Radiomic Features
Radiomic features are high-level quantitative features that can be extracted from images through mathematical operations. Radiomic features can be categorized into three subtypes: (i) intensity features, (ii) shape features, (iii) texture features. Intensity features are also known as first order features, which describe the histogram distribution of pixel/voxel values. Shape features describe the geometry of the Region of Interest (ROI) regardless of the intensity of the ROI. Texture features are based on the spatial distribution of the pixel/voxel values. Both MATLAB and Python have built-in libraries that support automatic extraction of large amount of radiomic features:
• MATLAB:Get Started with Radiomics - MATLAB & Simulink - MathWorks Australia
• Python:https://pyradiomics.readthedocs.io/en/latest/index.html#
For the potential use in classification, it is encouraged to extract all radiomic features supported by the library used.
Step Four: Feature Selection
Three properties are generally used for featureselection: repeatability, saliency, and compactness. In this project, we use repeatability as the criteria of feature selection. Conventional features are typically highly repeatable; however, the repeatability of radiomic features is not guaranteed.
Design a strategy for repeatability test; you can use the following case to understand the feature repeatability and as a guide to ‘mimic’ the differences between the two MRI examinations:
‘A patient with glioma had a brain MRI examination on Day 1 at Sir Charles Gairdner Hospital; on Day 2, the patient had another brain MRI examination at Fiona Stanley Hospital. Ignoring any tumor growth between Day 1 and Day 2, a feature is repeatable if when extracted from the two MRI examinations, the results are equal or equivalent.’
Select the top 10 intensity features, shape features, and texture features based on their repeatability.
Step Five: Classification using SVM
After the feature selection process, students will apply a Support Vector Machine (SVM) to classify the extracted features into categories of Low-Grade Gliomas (LGG) and High-Grade Gliomas (HGG).
Data Partition
Assign 10 LGG patients and 10 HGG patients to a ‘hidden’ testing set. Train and validate the SVM classifier on the rest of the dataset. Test the accuracy of classification using the ‘hidden’ testing set.
For data split between the training and the validation sets, you can consider either:
• Using a fixed data split, or;
• Using cross-validation.
Before data partition, find out the number of LGG/HGG patients. Note the huge class imbalance, which should be addressed.
The Effectiveness of SVM Classification
The effectiveness of the SVM in accurately classifying the glioma grades based on the selected features will be evaluated using accuracy:
Accuracy = num_of_correct_classifications / num_of_total_classifications
Deliverables
The deliverables include:
• A matlabor python GUI. The GUI should include the following components:
1. A ‘Load Slice Directory’ button, a ‘Channel’ drop-down menu, an ‘Annotation’ drop-down menu, and a ‘Slice ID’ slider.
a. Clicking the ‘Load Slice Directory’ should allow selecting a directory, which is a subfolder containing 155 H5 files belonging to the same MRI volume. You can assume the filenames and the data structure of the H5 files follow the convention of the downloaded dataset; however, the spatial resolution of the slices may vary from the downloaded dataset.
b. The ‘Channel’ drop-down menu should control the channel of the MRI volume displayed: selecting T1, T1Gd, T2, or T2-FLAIR should display the corresponding channel.
c. The ‘Annotation’ drop-down menu should control the annotation of the tumor displayed: selecting ‘On’ should superimpose the tumor mask on the MRI slice using an alpha value of 0.5; selecting ‘Off’ should not display the tumor mask.
d. The ‘Slice ID’ slider should control the slice of the volume visualized, dragging the slider should allow changing the slice displayed seamlessly.
2. An ‘Extract Conventional Features’ button.
Clicking on the ‘Extract Conventional Features’ button should allow selecting a directory containing multiple subfolders as described in 1.a. For each subfolder in the directory, the conventional features should be extracted. The output of ‘Extract Conventional Features’ should be a CSV file named ‘conventional_features.csv’ storing the extracted results, as shown in Fig. 3:
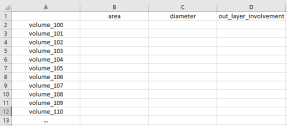
Figure 3 .
3. An ‘Extract Radiomic Features’ button.
Clicking on the ‘Extract Radiomic Features’ button should allow selecting a directory containing multiple subfolders as described in 1.a. For each subfolder in the directory, the top 10 intensity-based, shape-based, and texture-based radiomic features you selected based on repeatability should be extracted. The output of ‘Extract Radiomic Features’ should be a CSV file named ‘radiomic_features.csv’ storing the extracted results, as shown in Fig. 4:
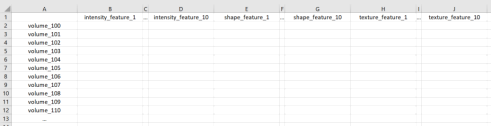
Figure 4. Note, the column names should be the names of the actual radiomic features selected.
• A live presentation during the lab sessions in week 12. The presentation should include (i) a display of the designed GUI; (ii) methods used for extracting the conventional features; (iii) strategy used for verifying the repeatability of the radiomic features; (iv) the top radiomic features selected according to repeatability; (v) the features used in the SVM classifier and the accuracy of classification during training and testing. Detailed schedule of the presentation will be released on LMS prior to week 12.
• A readme file detailing the SVM classification model. Importantly, the readme file should include:
• Detailed data partition;
• The features used for training;
• The accuracy of the model in classifying the MRI images during training, validation, and testing;
• A discussion of the SVM’s accuracy with regard to featureselection. Is using repeatability as the sole criteria for featureselection good?
• Any challenges encountered during the classification process.
Mark Distribution
• Visualization: GUI functional properly for ‘Load Slice Directory’, ‘Channel’, ‘Annotation’, and seamless slice-by-slice visualization. (15%)
• Conventional features: quality of the extraction of conventional features, which will be tested on a hidden set of MRI volumes. (35%)
• Repeatability test: the robustness and reasonableness of the designed repeatability test (15%)
• Radiomic features: repeatability of the selected radiomic features on a hidden set of MRI volumes. (25%)
• SVM Classification: accuracy and model discussion. (10%)